A British artificial intelligence company has shown that it can compute the shapes that proteins fold into, solving a 50-year problem in biology.
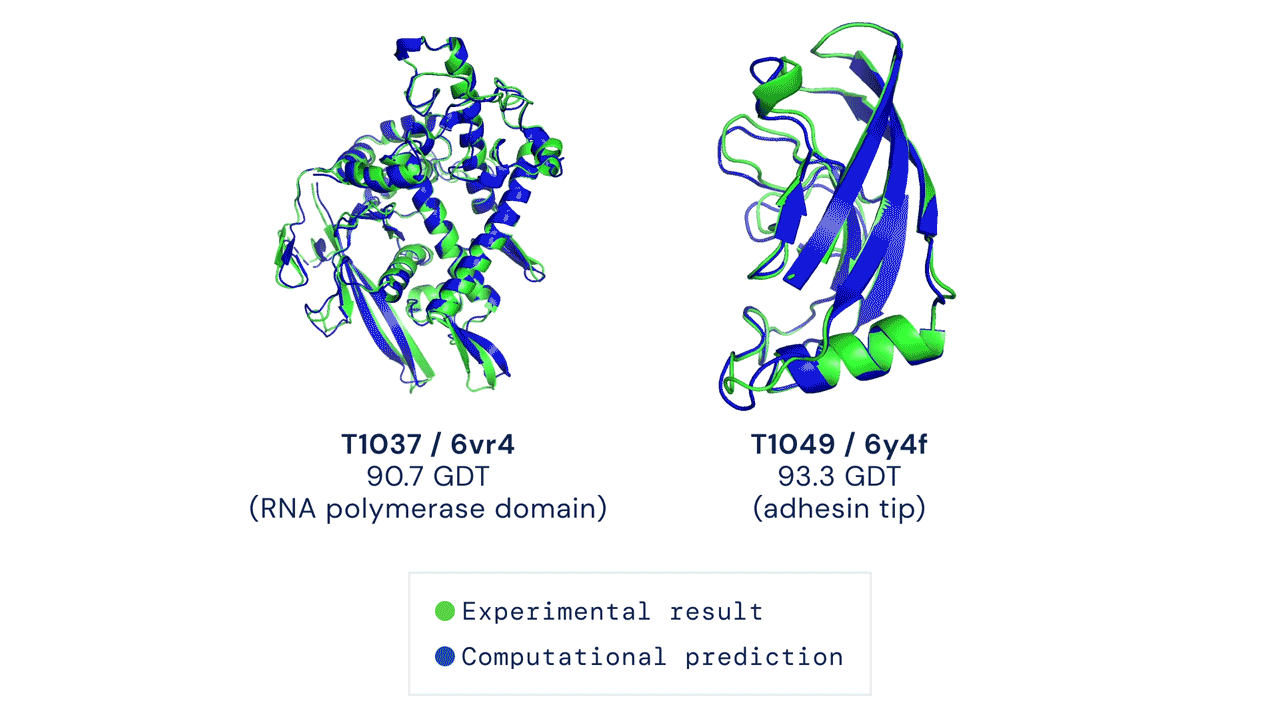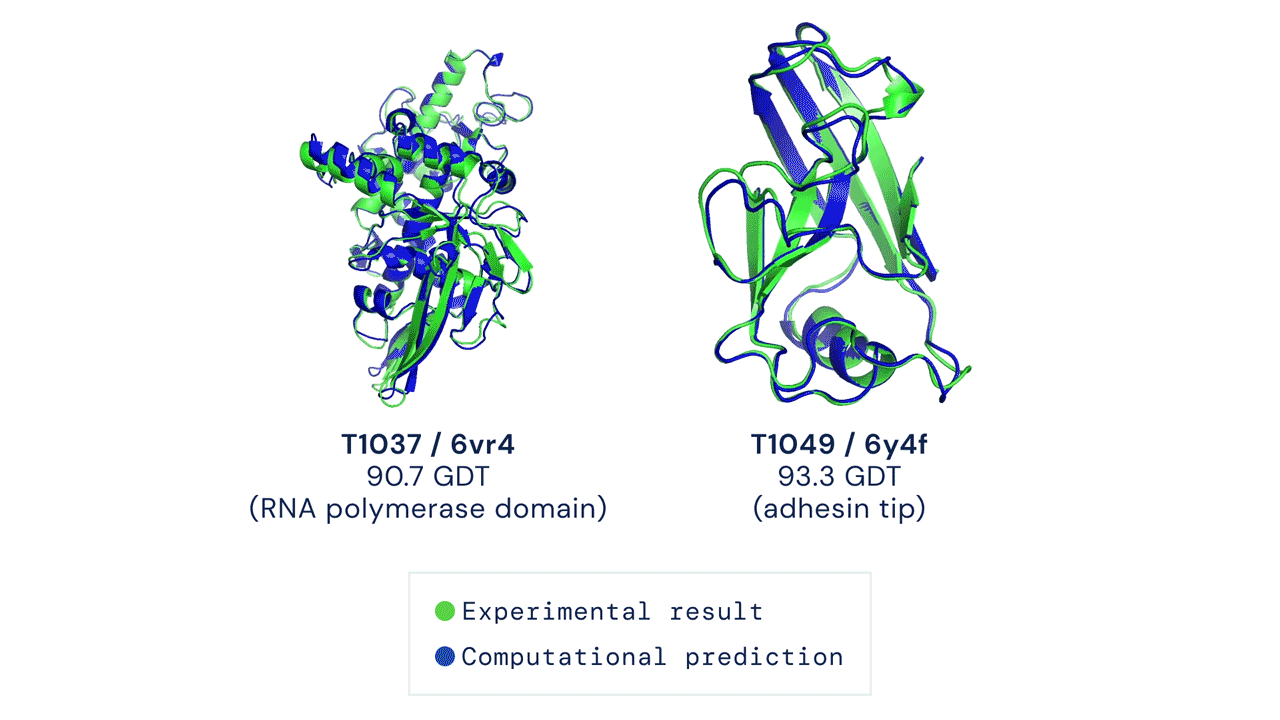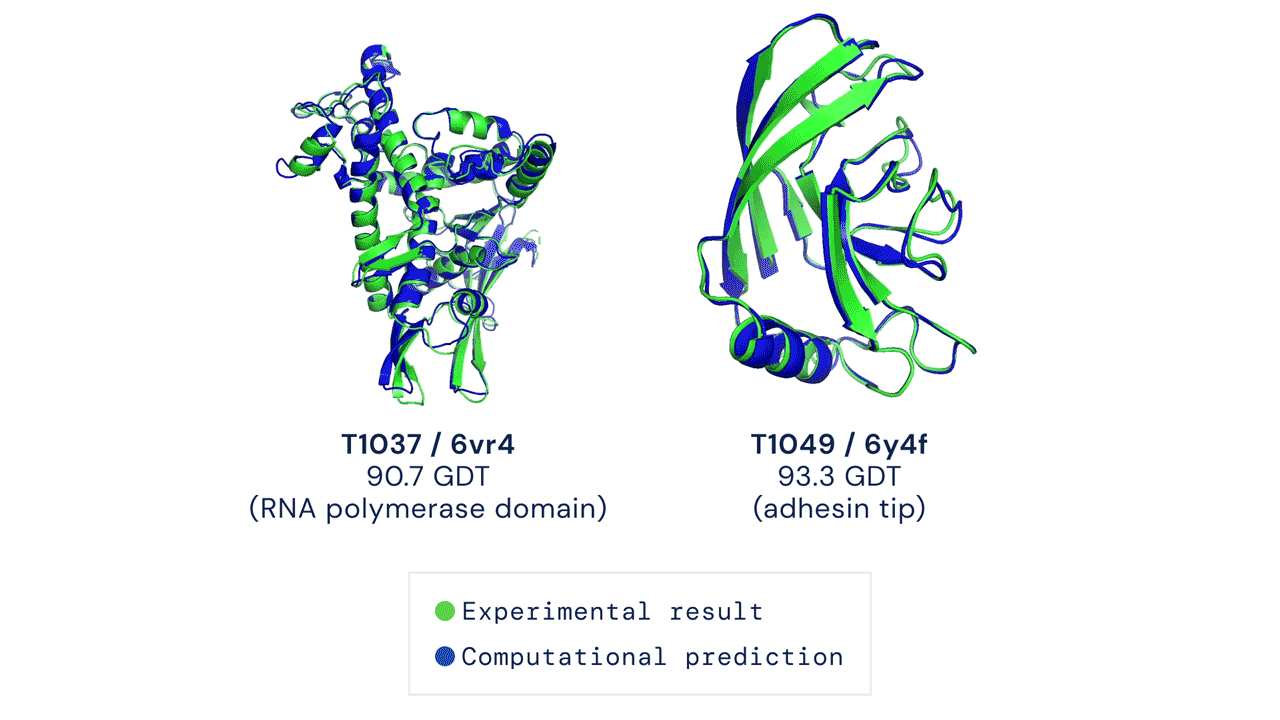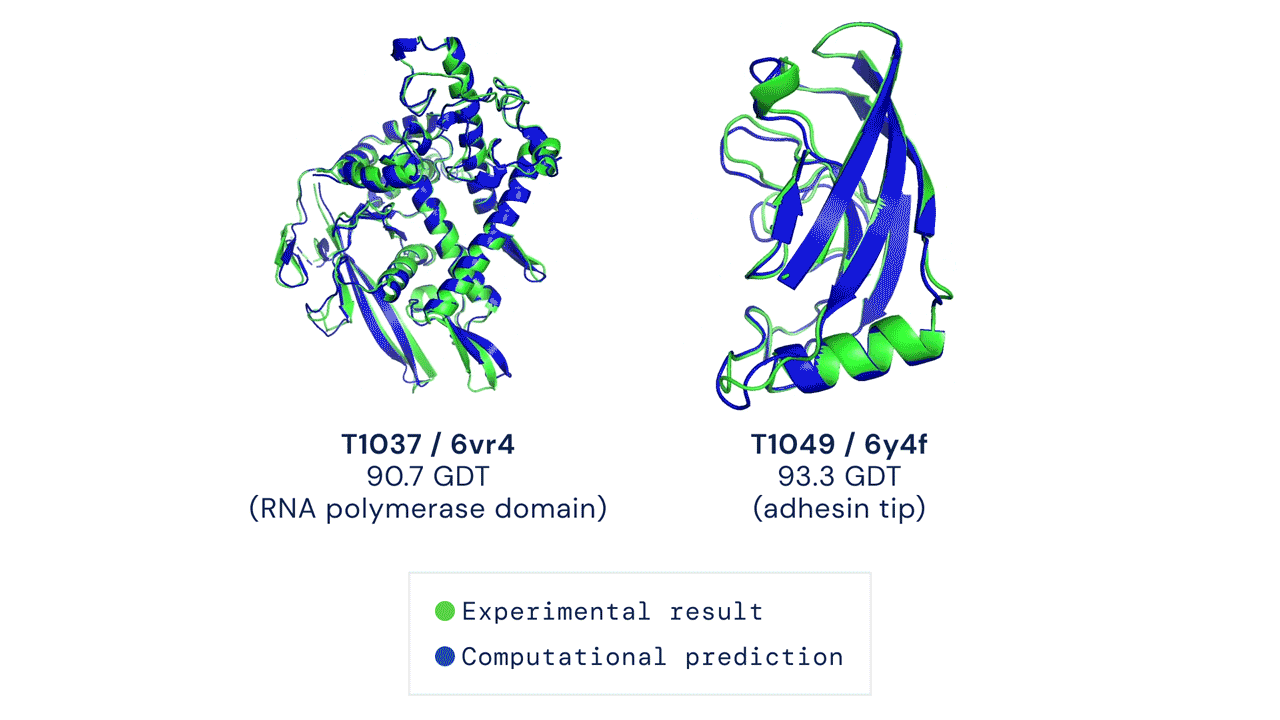
ABOVE: Two examples of protein targets in the free modelling category. AlphaFold predicts highly accurate structures measured against experimental result.
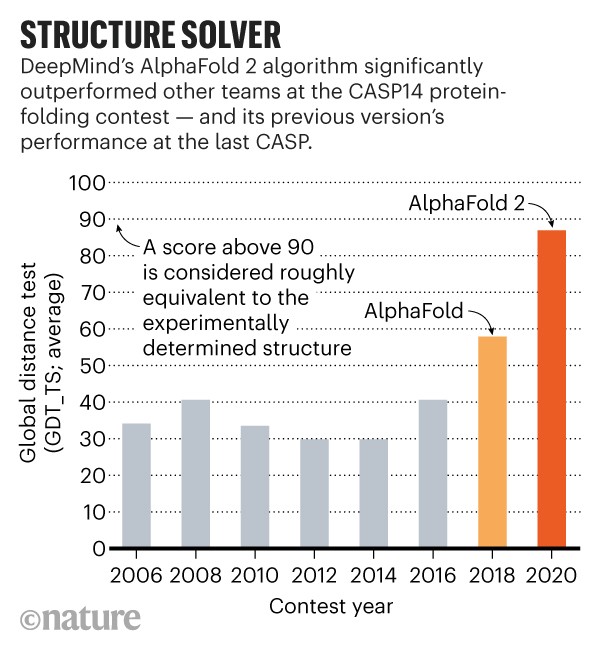
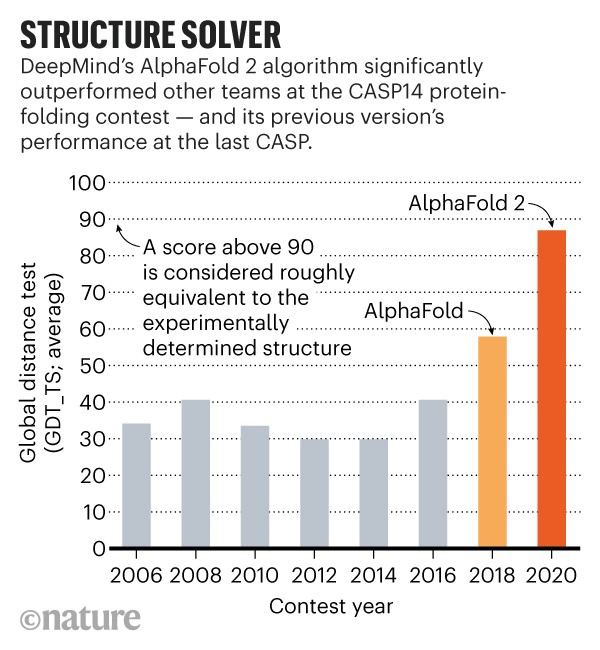
BELOW: AlphaFold is a once in a generation advance, predicting protein structures with incredible speed and precision. This leap forward … from AlphaFold to AlphaFold 2 … demonstrates how computational methods are poised to transform research in biology and hold much promise for accelerating the drug discovery process.
1 December 2020 (Chania, Crete) – As the story is told, it was a cold March day in 2016 and DeepMind CEO Demis Hassabis reportedly told computer scientist David Silver: “I’m telling you, we can solve protein folding.” Four years later, the London-based AI lab says they’ve figured out the protein-folding problem – a biological mystery that’s flummoxed scientists for 50 years.
On Monday, DeepMind announced a breakthrough in the area of protein folding, which uses a protein’s DNA sequence to predict its three-dimensional structure. With this new ability, we can better understand the building blocks of cells and enable advanced drug discovery. This is a huge scientific and industrial milestone. As Dr. Chris Donegan told me “this is a problem that has consumed computational biologists for decades. It is a real inflection point”.
As I noted in a long post last night to my science listserv, it’s really hard to overstate the importance of this, though it will take years to become apparent. It’s rather like how the sequencing of the human genome in the late 1990s led to us being able to produce an mRNA vaccine in just two days, given its sequence (the rest of the time has been spent on production and trials).
The complexity of protein folding is mindblowing: it depends on interactions between existing parts of the protein as it’s produced, which then are influenced by subsequent parts. And it all works 😳 . For those of us that have had the time to peer our complexity lens on the COVID-19 pandemic, it’s all even more astonishing. And thrilling. Between 2018 and 2020, Alphafold has gone from impressive to superhuman – and proving that machine learning approaches can solve the folding problem.
What DeepMind did, in a nutshell
I have followed DeepMind for almost 5 years. I got hooked in 2016 when I was in Zurich, Switzerland with my fellow artificial intelligence students. We got up early to watch the live feed from Korea as Lee Se-dol, a South Korean professional Go player, and AlphaGo, a computer program developed by DeepMind, played the board game Go. As we all know, Lee got trashed. I was fascinated. I even learned to play (rudimentary) Go. One of my AI professors made an introduction for me at DeepMind and I was off to a deep dive into their work.
My second degree at university was physics but over the last 20 years my primary science reading has been biology (I lost both parents to debilitating diseases) so I have a pretty good understanding of what DeepMind accomplished. To keep it simple, mitochondria may be *the powerhouse of the cell* but proteins are one of its main building blocks. So where does protein folding come in? Whichever shape a protein folds into will decide its function. The challenge is that proteins can take a virtually infinite number of forms. Bottom line: If you can predict a protein’s shape, then you can essentially predict its function. Since research suggests many diseases – Alzheimer’s, Parkinson’s, diabetes, cancer – are influenced by incorrect protein folding, this breakthrough is a really big deal for disease research and drug discovery.
Two years ago, the company released its first attempted protein-folding solution. But it wasn’t quite effective enough for use in the field. John Jumper, the protein-folding team’s senior researcher, said at the time:
“We used relatively off-the-shelf neural network technology [V1]. When the team tried to build on that version, they hit a real wall in what they were able to do.”
So this time, the team custom-built a neural network from the ground up. Unlike V1, which was made of separate components, the new AlphaFold is an end-to-end system – which can translate to more accurate results. The flip side: like most end-to-end systems, AlphaFold’s decision-making is opaque and hard to explain. While DeepMind can’t offer full insight into how the model arrived at a prediction, it will provide scientists with a confidence measure for each prediction. And a very detailed paper will be submitted for peer review.
And, yes, the critics have circled. Some critics say that due to lack of precision, it’s “laughable” to call the problem solved. Others will likely remain skeptical until they can examine AlphaFold’s code, calling on DeepMind to release it to the public domain. And virtually all plan to wait and see how the model performs IRL. For those of you who want more, the DeepMind briefing note is well worth your time. Just click here.
For biology buffs: Can’t get enough of protein folding? DeepMind created an illustrated video animation explaining how the process works:
THE BIG PICTURE: caveats aside, all the experts I spoke with agree this is a major step forward in disease research and drug discovery. This could an “Imagenet moment” that will trigger a wave of investment and innovation. Back in 2011, computer vision was stuck in the doldrums, not really good enough to generalise and be present everywhere. Then in 2012, Alex Krishevsky developed a model using convolutional neural networks that improved computer vision performance hugely. This triggered not just a boom in computer vision but a wave of venture and corporate investment in machine learning more widely. I’d expect AlphaFold 2 to do the same.
I watched the presentation on the BBC and I was struck by DeepMind’s founder Demis Hassabis:
It marks an exciting moment for the field, where it shows that these algorithms are becoming mature and powerful enough to become applicable to really challenging scientific problems.
It took Deepmind 10 years to get to this point. So I turned to long-time colleague Ben Blume (who has tracked DeepMind developments for years and who is a Principal at Atomico) to get his thoughts.
NOTE: Atomico is a European venture capital firm headquartered in London, with offices in Paris, Beijing, São Paulo, Stockholm, and Tokyo. They also publish every year their “State of European Tech” report, in partnership with Slush and the U.S. law firm Orrick Herrington. It is considered the definitive report on pretty much every aspect of European tech. And the Slush technology conference/investor “meet & greet”/tech workshop event, held in Helsinki, Finland, is where I would always end my tech conference gauntlet every year, at least before I retired. Despite the sub-zero temperatures, more than 25,000 people usually show up. It is at Slush that Atomico formally presents its “State of European Tech” report with a 1.5 hour discussion of the report.
In brief, DeepMind Technologies is a UK based artificial intelligence company and research laboratory founded in September 2010, and acquired by Google in 2014. The company is based in London, with research centres in Canada, France, and the United States.
The company first hit the media when it created a neural network that learned how to play video games in a fashion similar to that of humans (see below), as well as a Neural Turing machine, or a neural network that may be able to access an external memory like a conventional Turing machine, resulting in a computer that mimics the short-term memory of the human brain.
But it hit the big time in 2016 after its AlphaGo program beat a human professional Go player Lee Sedol, the world champion, in a five-game match.
NOTE: I was in Zurich for several days when those games were played and we watched them live. You can read my report and analysis by clicking here.
So let’s take a short trip down memory lane with Ben as he hits the high points of DeepMind history:
2013: Playing Atari with Deep Reinforcement Learning. A simple problem in a well constrained environment, outperforming a human, but ‘beating’ a computer intelligence. Enough to capture the imagination about what might be was possible. More here.
2015-2016: AlphaGo. From the first publicized win against the reigning European Champion to the famous victory over Lee Sedol, this was highly publicised proof that AI could beat a human intelligence, and was developing faster than many experts expected. More here.
2016: WaveNet. Taking on a different aspect of ML, WaveNet was a radical improvement in decades old Text-to-Speech tech. The Duplex demo 18 months later moved this tech into the sci-fi realm and made people think about how fast AI was moving into their lives. More here.
2016: Data Centre Cooling. Another well constrained problem, this always impressed me for the practical real world nature of the task, and the significant degree of improvement in what was likely an already highly optimised system. More here.
2018: Streams. A collection of healthcare research inc “detecting eye disease more quickly and accurately; planning cancer radiotherapy treatment in seconds; and detecting patient deterioration from EHR”, Streams moved into problems with real world impact. More here.
2018: AlphaZero. A further iteration of the game playing work building from AlphaGo, I love the idea that both Lee Sedol and Kasparov felt the AI had developed its own unique style that they could take inspiration from. AI teaching humans. More here.
2019: AlphaStar. Significant here was how much more complex the environment was than any of the previous work. No optimum strategy, imperfect information, real time, huge action space. AlphaStar didn’t take long to reach Grandmaster level. More here.
2018-2020: AlphaFold. “Algorithms mature enough to solve really challenging scientific problems”. Amazing to see such radical improvement on many decades of research. More here.
What a mind-blowing contribution in just 10 years. Amazing to think what is next.